Introduction
You already saw this in spellcheckers: a list with correct words which are similar to the misspelled word you've typed. Wouldn’t it be nice to incorporate this notion of similar words as a functionality in your applications? There are lots of possible uses for it:
- The most obvious case is searching for a name in a database. People with an unusual name always need to patiently spell it and hope it is written correctly in the database. Now, imagine you could scan a database and search for names that are similar (look alike) to any given name and rank them by similarity, so the user could choose them. Cool, eh?
- Show a list of web addresses similar to the one the user typed wrongly and was not found. Or use it in a search for "approximate" keywords in your site.
- You may also use this idea to check similarities between written text (academic papers, source code of programs, etc.) to detect plagiarism, for instance.
- Hey, what about comparing genetic sequences, and to discover evolutionary and functional similarities among genes or proteins? Actually, this was done with mitochondrial DNA to trace all the human ethnical evolution and create the concept of Mitochondrial Eve… OK, OK, insanely off-topic...
Here, I offer a possible solution for implementing such comparisons: a function that returns how much two strings are “alike” or similar. But first, let’s define the problem more precisely.
Background
Similarity of strings can be defined in more than one way. Here, I will use a very simple one: the length of the longest common sequence divided by the length of the first string. Suppose you have two strings (e.g.: “consecutivelly” and “successfully”). Now see that you have several sub sequences that appear in both strings, but not necessarily consecutively (e.g.: “c-s-u-l-l-y”, “s-c-u-l-l-y”, “s-e-u-l-l-y”, "s-u-e-l-l-y", etc.). The measure of similarity I propose is the division of the length of the longest of these subsequences by the length of the first parameter string (e.g., strlen(“suelly”)/strlen(“consecutively”), which is approximately 42.85%). So far this is simple, right?
Now, the hard part seems to be how to discover efficiently that longest subsequence. Well, actually this is not so hard: we can use a classical algorithmic technique called dynamic programming. If you studied algorithms in any university and they didn’t teach you it, ask your money back. Very grossly, you can think of dynamic programming (or DP) as a kind of reversed recursion used in optimization problems. Some optimization problems have a recursive solution. However, this recursive solution may involve the computation of some sub solutions which can appear repeated times. To such problems, we can use DP with great gains of performance. It would be too lengthy to explain here this technique in detail, so I strongly suggest you to take a look at a good book on algorithms (I suggest one at the bottom in the Points of Interest section). By the way, “programming” in “dynamic programming” doesn’t refer to computer programming but to programming in a general mathematical sense; i.e.: a sequence of steps which lead to a solution.
Using the code
The code is very simple to use. The function which returns the similarity has the following signature (in string.h):
float simil( char *str1, char *str2);
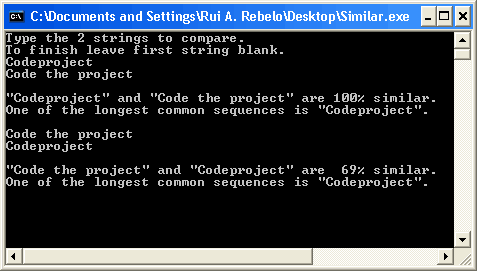
The return value is a number between 0 and 1. This value, as I mentioned before, is the length of the longest common sequence divided by the length of str1. 0 means the two strings have nothing in common. 1 means the first string is totally contained in the second. It is up to you to decide which string to use as first or second. But please take notice that using the longest string as the first parameter yields lower similarity values and using the shortest yields higher (look at the picture above). Obviously, the decision on how you make the call and which value will be the minimum acceptable will depend on the problem you are trying to solve. If you are scanning names in a database, you might want to let the user adjust this value. If you are building a script for a 404 response page in a website then you will adjust this value comparing the most common errors to a list of addresses in your site.
I also provide another function you might be interested in:
char *LCS( char *str1, char *str2);
This function returns a pointer to a static array of char (with maximum size MAX_LCS, defined in string.h) containing one of the longest sequences. All you have to do is to compile and link simil.c to your code and leave simil.h in a folder in your include path.
Points of Interest
I made this code when studying a more generic problem called edit distance, which is a measure of how much a string (or DNA sequence, throughout evolution and by genetic mutations) must be changed to generate another one. This is a typical problem for dynamic programming. You can read a deep and complete explanation of what is dynamic programming in this book: Introduction to Algorithms, 2nd edition; by Thomas H. Cormen, Charles E. Leiserson, Ronald L. Rivest and Clifford Stein. I have a love/hate relationship with this book. It is one of the two most complete books on algorithms I've ever seen (the other is Knuth's). But it is also one of the most pedantic books I've ever read. It is the typical academic book: deep knowledge, lots of proofs, and convoluted formalisms. Robert Sedgewick's "Algorithms in C" also has an introduction to DP, but not so complete. If you have another suggestion of book or site on dynamic programming, please post it in the comments. :-)
History
- 12/September/2004: Version 1.0.
 General
General  News
News  Suggestion
Suggestion  Question
Question  Bug
Bug  Answer
Answer  Joke
Joke  Praise
Praise  Rant
Rant  Admin
Admin 







 But I think the language is powerful enough to support what my algorithm does.
But I think the language is powerful enough to support what my algorithm does. 


 I was afraid of hearing that. Dynamic Programming is not easy to explain, especially in a short web article. But I'll try.
I was afraid of hearing that. Dynamic Programming is not easy to explain, especially in a short web article. But I'll try.